Classical molecular dynamics (MD) is a computational technique that provides conformations of molecular/atomistic systems compatible with given thermodynamic ensembles.
Enhanced sampling techniques are particular methods that improve the sampling capabilities of plain MD and force the exploration of regions of conformational space that could be relevant, but difficult to achieve in a regular simulation.
- We use enchanced sampling molecular dynamics to force opening of potential binding and ligand-interaction sites on the proteins' surface.
- We trigger this opening by tweaking the behavior of water molecules.
Using combined computational and experimental approach we investigate the effects of mutations on the function of proteins affected in rare genetic diseases.
- Within the project, our group is involved in enhanced sampling MD simulations exploring the chloride permeation pathway, and developing software aimed at predicting the effects of CLC mutations.
- Collaboration:



- Project funding:

- Within the STOP-SPREAD Bad Bugs consortium, we conduct computational and experimental analyses of antibacterial peptides and small molecules interacting with bacterial membranes. We work at the interface of academia and industry, in collaboration between IIT and an industrial partner.
- Collaboration:

Project funding:
Completed projects
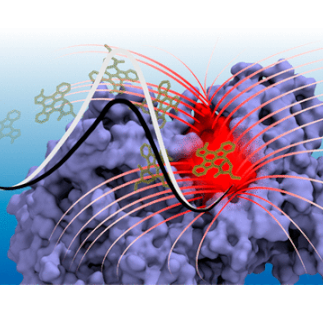
Protein-ligand unbinding protocol
Luca Mollica, Sergio Decherchi, Syeda Rehana Zia, Roberto Gaspari, Andrea Cavalli and Walter Rocchia; Kinetics of protein-ligand unbinding via smoothed potential molecular dynamics simulations. Scientific Reports 5, 2015
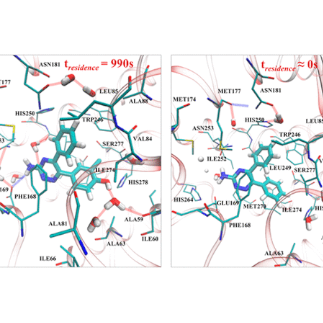
Docking Guided by Adaptive Electrostatic Bias
A. Spitaleri, S. Decherchi, A. Cavalli and W. Rocchia; Fast Dynamic Docking Guided by Adaptive Electrostatic Bias: The MD-Binding Approach. Journal of Chemical Theory and Computation 14(3) 2018
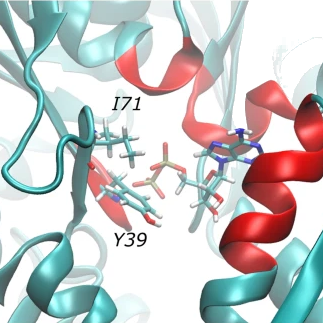
Dynamical characterization of water persistency in binding sites
S. Rehana Zia, R. Gaspari, S. Decherchi and W. Rocchia. Probing Hydration Patterns in Class-A GPCRs via Biased MD: The A2A Receptor. Journal of Chemical Theory and Computation, 12(12), 2016